The Abyss of Genetic Complexity and Lifestyle
Polygenic diseases are a very interesting and complex area of genetics where multiple genes are involved.
Unlike monogenic diseases where a single gene mutation causes the disease, polygenic diseases are caused by many gene mutations interacting with each other to contribute to the onset of the disease.
This page focuses on lifestyle-related diseases such as hypertension and diabetes, and provides a detailed explanation of advanced genetic research techniques such as genome-wide association studies (GWAS), single nucleotide polymorphisms (SNPs), linkage analysis, and Mendelian randomization.
Understanding Polygenic Disorders
Common examples of polygenic diseases include cardiovascular disease, diabetes, obesity, depression, and schizophrenia.
These diseases are considered multifactorial because they are influenced by both genetic and environmental factors, and unlike monogenic diseases, they are caused by the cumulative effect of many genes.
Lifestyle-related diseases: High blood pressure and diabetes
[High Blood Pressure]
Hypertension is a chronic condition characterized by persistently high blood pressure in the arteries. It is a major risk factor for cardiovascular diseases such as heart attack and stroke.
- Genetic factors
- Environmental factors
Many genetic loci are associated with hypertension, including genes that regulate physiological processes such as renal sodium handling, vascular tone, and the renin-angiotensin system.
Lifestyle habits such as a high-salt diet, lack of exercise, and stress significantly affect the risk of high blood pressure.
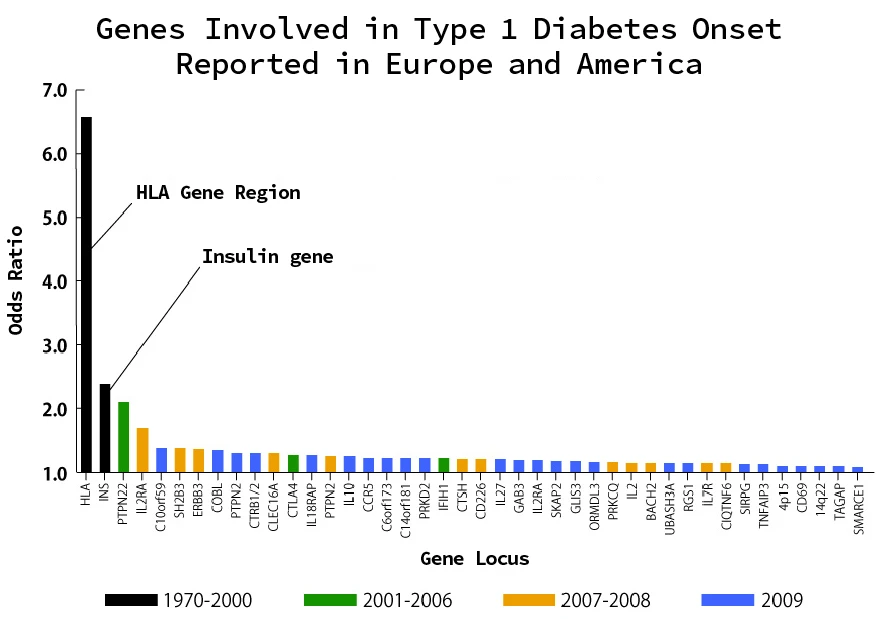
[Diabetes mellitus]
Diabetes, particularly type 2 diabetes, is a metabolic disease characterized by hyperglycemic states due to insulin resistance or insufficient insulin secretion.
- Genetic factors
- Environmental factors
Several genetic variants that affect insulin secretion and glucose metabolism have been identified, often in genes involved in pancreatic β-cell function and the insulin signaling pathway.
Diet, obesity, and lack of exercise are important environmental factors in the development of type 2 diabetes.
Single nucleotide polymorphisms (SNPs)
[What is a SNP?]
Single Nucleotide Polymorphism (SNP) is the most common form of genetic variation in genes and refers to a difference in a single base (A, T, C, G) that makes up DNA at a specific position in the genome. The human genome contains about 3 billion base pairs of DNA, among which about 10 million SNPs are known. These variations are an important factor in forming genetic diversity between individuals.
- ●How to detect SNPs
- ①DNA sequencing
- ②Genotyping array
- ●SNP database
- ①dbSNP
- ②1000 Genomes Project
Technologies for detecting SNPs are evolving rapidly. Common methods include.
Next-generation sequencing (NGS) technology is used for fast and accurate detection of SNPs across large genomes.
Specific SNP-targeted array technologies are used to detect many SNPs at once, and these arrays are cost-effective and suitable for large population studies.
The detected SNPs are registered in databases. Representative databases include the following.
This is a database of SNPs provided by NCBI, and contains information on millions of SNPs.
This project comprehensively covers human genetic diversity and makes SNP data collected from different populations around the world publicly available.
[Classification of SNPs]
SNPs are classified based on the gene function at their location as follows.
- ①Coding region SNPs
- Synonymous SNPs
- Nonsynonymous SNPs
- ②Non-coding region SNPs
A mutation that does not affect the amino acid sequence coded by a gene. The encoded amino acids are coded for by three bases (codons), which code for 20 different amino acids. Therefore, there are multiple combinations of bases that code for one amino acid. For example, the codons GAA and GAG both code for glutamic acid. Therefore, even if the third base mutates from A to G, the same amino acid is coded for. Therefore, there is no difference in the coded amino acid sequence.
A mutation that changes the amino acid sequence encoded by a gene. Nonsynonymous SNPs are further divided into missense mutations (where one amino acid coded for by a gene is replaced by another) and nonsense mutations (where an amino acid is replaced by a stop codon, terminating the encoded amino acid sequence).
Mutations located in regions that regulate the location and timing of gene expression (promoters and enhancers) or intron regions.
They can affect gene splicing and the efficiency of transcription.
[Functional effects of SNPs]
SNPs have various effects on the functions of genes and proteins, including the following.
- ①Regulation of gene expression
- ②Changes in protein function
- ③Changes in splicing
SNPs located in regulatory regions can alter gene expression levels through transcription factor binding and changes in chromatin structure.
Nonsynonymous SNPs alter the amino acid sequence of a protein, thereby affecting its function and stability.
SNPs located at intron-exon boundaries can alter splicing patterns.
[SNP research and applications]
- ①Disease-related research
- GWAS
- Disease risk prediction
- Functional studies
- ②Personalized medicine
- ③Evolution and population genetics
Research to identify genetic variants associated with disease using SNPs. Finding the association between a specific SNP and a disease clarifies the onset mechanism and risk factors of the disease. GWAS will be explained in more detail in the next section.
When the presence of a particular SNP is known to increase the risk of a particular disease, for example through GWAS results, it can be used to assess an individual's risk.
Investigating how differences in specific SNPs affect gene function can be used to deepen our understanding of disease pathology and elucidate the mechanisms underlying disease development.
Using SNP information, it is possible to select the most appropriate treatment and drug for each individual patient. For example, SNPs in genes involved in drug metabolism can affect the efficacy and side effects of drugs, helping to determine the appropriate dosage for each patient.
It also impacts risk assessment and progression prediction for polygenic diseases.
SNP data are used to study genetic variation among different populations,which can shed light on human evolution, migration patterns, and genetic relationships between populations.
Genome-wide association study (GWAS)
GWAS are studies that scan the entire genome for SNPs associated with a particular disease or trait. By comparing the genomes of people with and without the disease, genetic variants that contribute to disease risk can be identified.
GWAS collects DNA samples from large cohorts and analyzes hundreds of thousands to millions of SNPs using high-throughput genotyping.
As a result, many risk loci related to hypertension and diabetes have been identified by GWAS. Although the contribution of each locus identified by GWAS to the disease is small, their accumulation contributes to the development of polygenic diseases.
Linkage analysis
Linkage analysis is a method to create a genetic map and identify disease-causing genes by examining the inheritance patterns of marker genes (SNPs) within families.
- ●Principle
- ●Application
During meiosis, which produces sperm and eggs, genetic recombination occurs between homologous chromosomes.
The frequency of recombination of genes that are physically close to each other is lower than that of genes that are far apart, making them more likely to be inherited together.
Based on this frequency, the distance between genes is estimated and a genetic map is created.
From there, marker genes that are closer (more likely to be inherited together) are found to narrow down the causative gene, and ultimately the causative gene is identified.
Although linkage analysis is primarily used for monogenic diseases, it can also be useful for identifying genomic regions that contribute to polygenic traits.
However, due to the increased complexity of the analysis, it is not as effective as GWAS for diseases with complex inheritance patterns, such as polygenic diseases.
Mendelian randomization
Mendelian randomization is a method that uses genetic variants (SNPs) to infer causal relationships between risk factors and disease in observational studies.
- ●Concept
- ●Application
When investigating the effectiveness of a drug, patients can be randomly assigned to either a group that receives the drug or a group that does not, allowing the causal relationship between the drug and the treatment outcome to be determined.
However, polygenic diseases are influenced by both environmental and genetic factors, making it difficult to determine the causal relationship between a specific risk factor and the disease.
Therefore, we use the fact that genes are randomly distributed from parents to children to estimate the causal relationship between risk factors and the disease.
This mimics the randomization process of clinical trials.
It helps to clarify whether the associations seen in observational studies are causal.
For example, let's say you want to estimate the causal relationship of high body mass index (BMI) causing diabetes. First, find a gene (SNP) that is not related to diabetes but is correlated with BMI. You can estimate the causal relationship by evaluating the proportion of people with this gene and the proportion of people with diabetes.
An integrated approach
Modern research combines SNP analysis, GWAS, linkage analysis, and Mendelian randomization to provide a comprehensive understanding of polygenic diseases.
These integrated approaches help elucidate the genetic architecture of diseases and identify potential therapeutic targets,thereby aiming to target causal rather than symptomatic treatments.
Summary
Lifestyle-related diseases such as hypertension and diabetes are polygenic diseases that represent a complex interplay between genes and the environment.
Advanced genetic research techniques such as SNP analysis, GWAS, linkage analysis, and Mendelian randomization are powerful tools to unravel this complexity.
Understanding these diseases at the molecular level opens the door to personalized medicine based on an individual's genetic makeup.
Exploring the depths of polygenic disease helps us understand the incredible complexity of human health and disease, and highlights the need for continued research and innovation in genetic and environmental factors.